Metadata defines the NetCDF format
Network Common Data Form (NetCDF) is the arguably the most popular data format in the field of atmospheric sciences, meteorology and oceanography. To quote the description of this format by the official site:
Data in netCDF format is:
Self-Describing. A netCDF file includes information about the data it contains.
Portable. A netCDF file can be accessed by computers with different ways of storing integers, characters, and floating-point numbers.
Scalable. Small subsets of large datasets in various formats may be accessed efficiently through netCDF interfaces, even from remote servers.
Appendable. Data may be appended to a properly structured netCDF file without copying the dataset or redefining its structure.
Sharable. One writer and multiple readers may simultaneously access the same netCDF file.
Archivable. Access to all earlier forms of netCDF data will be supported by current and future versions of the software.
The 1st defining attribute of the NetCDF format is identified as Self-describing, and I believe it is also the most important one. NetCDF files contain not only data, but also metadata that describe what the data are about, the name, units, data type, and data creators etc.. Anyone with some suitable tools should be able to get at least some basic understandings of the data just by reading the metadata.
There exist libraries to access the NetCDF format in C, Fortran, C++, Python, and other languages. For Python, the relevant library is called netcdf4. On top of that, there are also some other packages that offer more advanced functionalities, for instance CDAT, iris and xarray. I’m still on my way learning xarray, and have had little experience with iris, so the comparison will be centered around CDAT and netcdf4.
Sample data
We will be using some sample NetCDF data as an illustration. You can obtain the zipped data file from here. The data is the monthly average global Sea Surface Temperature (SST) of year 1989, from the ERA-Interim reanalysis dataset. Below lists some basic information about the file:
- File name:
sst_s_m_1989_ori-preprocessed.nc - File size: ~ 2.1 Mb
- Data rank: 4
- Data dimensions: 12 (time) x 1 (level) x 241 (latitude) x 480 (longitude)
- Data id: ‘sst’
- Data unit: ‘K’
- Data long_name: ‘Sea surface temperature’
- Horizontal resolution: 0.75 x 0.75 degree latitude/longitude
- Temporal resolution: monthly
The data are really not important here, you can follow along the article without using any actual data.
Read NetCDF data using CDAT
Below is the code snippet to read in the data using the cdms2 module of the CDAT package:
import cdms2 as cdms
abpath_in=os.path.join(SOURCEDIR, 'sst_s_m_1989_ori-preprocessed.nc')
print('\n### <netcdf_manipulation>: Read in file:\n',abpath_in)
fin=cdms.open(abpath_in,'r')
var_cdat=fin('sst')
fin.close()
SOURCEDIR is the path to the folder where you saved the sample data file.
We assigned the read-in data to the var_cdat variable, which, by the definition of NetCDF format, should be a combination of data values and metadata. Let’s do some queries to check this out:
print('type of var_cdat =', type(var_cdat))
print('id = ', var_cdat.id)
print('standard_name = ', var_cdat.standard_name)
print('long_name = ', var_cdat.long_name)
print('unit = ', var_cdat.units)
print('shape = ', var_cdat.shape)
The output we get:
type of var_cdat = <class 'cdms2.tvariable.TransientVariable'> id = sst standard_name = Sea surface temperature long_name = Sea surface temperature unit = K shape = (12, 1, 241, 480)
So the var_cdat variable is a cdms2.tvariable.TransientVariable, which is a wrapper class built on top of the numpy.ma.core.MaskedArray class. Masked array facilitates manipulation of data with missing values, in this case, the land areas where SST has no definition.
The standard_name, long_name and units attributes tell us what the data is about and in what unit it is measured. The shape attribute tells the rank and orderings of the data array.
Now do some query on the coordinate system of the data. This is how you obtain the time and latitude axes of a TransientVariable:
timeax=var_cdat.getTime()
timeax_comp=timeax.asComponentTime()
print('timeax =')
print(timeax[:])
print('timeax_comp =')
print(timeax_comp)
print('id of time =', timeax.id)
print('unit of time =', timeax.units)
print('\n')
latax=var_cdat.getLatitude()
lonax=var_cdat.getLongitude()
print('id of latax =', latax.id)
print('unit of latax =', latax.units)
Below is the output:
timeax = [30680. 30711. 30740. 30771. 30801. 30832. 30862. 30893. 30924. 30954. 30985. 31015.] timeax_comp = [1984-1-1 0:0:0.0, 1984-2-1 0:0:0.0, 1984-3-1 0:0:0.0, 1984-4-1 0:0:0.0, 1984-5-1 0:0:0.0, 1984-6-1 0:0:0.0, 1984-7-1 0:0:0.0, 1984-8-1 0:0:0.0, 1984-9-1 0:0:0.0, 1984-10-1 0:0:0.0, 1984-11-1 0:0:0.0, 1984-12-1 0:0:0.0] id of time = time unit of time = days since 1900-1-1 id of latax = latitude unit of latax = degrees_north
Notice that we obtained the time axis/dimension (I’ll be using these two terms interchangeably) using
timeax = var_cdat.getTime()
timeax[:] shows the values of the time axis, which is an 1d array of floats. This is typically how the NetCDF file encodes time stamps: it uses a numerical array to save the amount of time lapses since a given reference time point. In this case, the reference is days since 1900-1-1 (with 00:00 UTC being implicit).
However, this is not particularly readable, so the asComponentTime() method does the translation work for you, and the output is
timeax_comp =
[1984-1-1 0:0:0.0, 1984-2-1 0:0:0.0, 1984-3-1 0:0:0.0, 1984-4-1 0:0:0.0, 1984-5-1 0:0:0.0, 1984-6-1 0:0:0.0, 1984-7-1 0:0:0.0, 1984-8-1 0:0:0.0, 1984-9-1 0:0:0.0, 1984-10-1 0:0:0.0, 1984-11-1 0:0:0.0, 1984-12-1 0:0:0.0]
This is much more human-readable. That is to say, 30680.0 days since 1900-01-01 00:00 UTC is 1984-1-1 0:0:0.0. There are also different types of calendars, but I’m not going to bother you with such nuances for now.
The latitude axis is obtained using var_cdat.getLatitude(), and it has an id of latitude, and a unit of degrees_north. We can also query the values of the latitude axis by latax[:], which again would be a numerical array. This is not shown for brevity.
Read NetCDF data using netcdf4
We repeat the similar reading and querying operations using the netcdf4 package, using the code shown below:
from netCDF4 import Dataset, num2date
abpath_in=os.path.join(SOURCEDIR, 'sst_s_m_1989_ori-preprocessed.nc')
print('\n### <netcdf_manipulation>: Read in file:\n',abpath_in)
fin=Dataset(abpath_in, 'r')
var_nc4=fin.variables['sst']
print('type of var_nc4 =', type(var_nc4))
print('name = ', var_nc4.name)
print('standard_name = ', var_nc4.standard_name)
print('long_name = ', var_nc4.long_name)
print('unit = ', var_nc4.units)
print('shape = ', var_nc4.shape)
print('\n')
timeax_nc4=fin.variables['time']
timeax_comp_nc4=num2date(timeax_nc4[:], timeax_nc4.units,
only_use_cftime_datetimes=False,
only_use_python_datetimes=True)
print('timeax =')
print(timeax_nc4[:])
print('timeax_comp =')
print(timeax_comp_nc4)
print('name of time =', timeax_nc4.name)
print('unit of time =', timeax_nc4.units)
print('\n')
latax_nc4=fin.variables['latitude']
print('name of latax =', latax_nc4.name)
print('unit of latax =', latax_nc4.units)
Notice that we obtained the time axis of the data not from the data variable var_nc4 itself, but from the file handle fin:
timeax_nc4=fin.variables['time']
And the time stamp translation work is achieved differently, using the num2date() function. Other than these, there is not much of a difference, the var_nc4 variable is also a combination of data values and metadata. Below is the outputs:
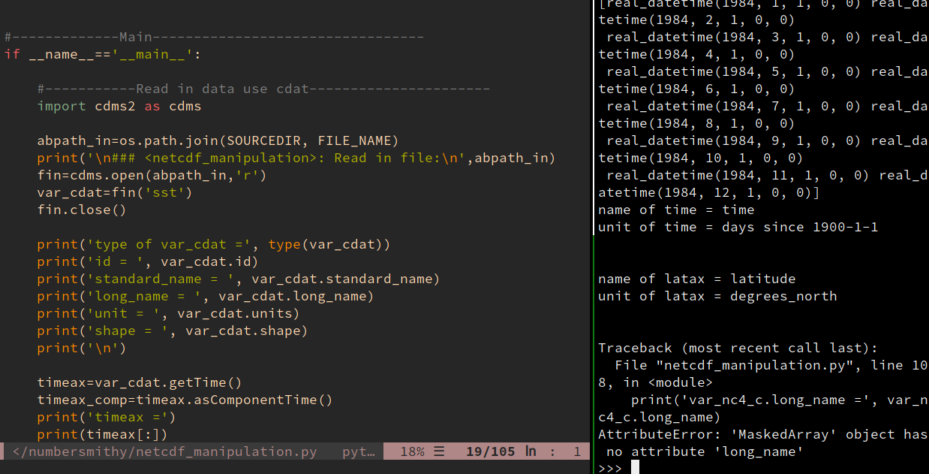
type of var_nc4 = <class 'netCDF4._netCDF4.Variable'> name = sst standard_name = Sea surface temperature long_name = Sea surface temperature unit = K shape = (12, 1, 241, 480) timeax = [30680. 30711. 30740. 30771. 30801. 30832. 30862. 30893. 30924. 30954. 30985. 31015.] timeax_comp = [real_datetime(1984, 1, 1, 0, 0) real_datetime(1984, 2, 1, 0, 0) real_datetime(1984, 3, 1, 0, 0) real_datetime(1984, 4, 1, 0, 0) real_datetime(1984, 5, 1, 0, 0) real_datetime(1984, 6, 1, 0, 0) real_datetime(1984, 7, 1, 0, 0) real_datetime(1984, 8, 1, 0, 0) real_datetime(1984, 9, 1, 0, 0) real_datetime(1984, 10, 1, 0, 0) real_datetime(1984, 11, 1, 0, 0) real_datetime(1984, 12, 1, 0, 0)] name of time = time unit of time = days since 1900-1-1 name of latax = latitude unit of latax = degrees_north
Difference between CDAT ant netcdf4
However, differences start to emerge as you go beyond such trivial attribute querying operations and perform some more complex manipulations of the data. For instance, let’s do some simple slicing of the data, by cutting out the domain of East Asia-Northwestern Pacific in the box of 10 - 60 degree North, and 80 - 150 degree East.
To do this with the var_cdat variable, it would be
var_cdat_c_sub=var_cdat_c(latitude=(10, 60), longitude=(80, 150))
print('var_cdat_c_sub.getLatitude() =', var_cdat_c_sub.getLatitude())
We sliced the data using the latitude/longitude coordinates, and queried the latitude axis of the subdomain. Here is the output:
var_cdat_c_sub.getLatitude() = id: latitude
Designated a latitude axis.
units: degrees_north
Length: 67
First: 10.5
Last: 60.0
Other axis attributes:
axis: Y
long_name: latitude
realtopology: linear
Python id: 0x7f59d74b2760
What if we want to derive a new variable out from var_cdat, say convert the units from K to Celsius. We can achieve this using:
var_cdat_c=var_cdat-273. var_cdat_c.units='C'
The new var_cdat_c is another TransientVariable, with all the same metadata as var_cdat, except that we need to update its units attribute.
Now let’s see how to do these operations with the var_nc4 variable.
Unfortunately, there is no built-in method to slice the data domain using dimension coordinates, like what we did with the var_cdat variable. For var_nc4, we will have to do the following steps:
- Obtain the latitude and longitude axes (which we already did).
- Locate the indices (
lat_idx1andlat_idx2) for the latitude bounds (10and60) from the latitude coordinates. And do the same for the longitude bounds to getlon_idx1andlon_idx2. - Slice the data value using the above obtained indices:
subdomain = var_nc4[:, :, lat_idx1:lat_idx2, lon_idx1:lon_idx2]
(Note this assumes you already knew where the latitude and longitude dimension are in the rank.)
-
If you also need the coordinate information for this new subdomain variable, you will have to slice the latitude and longitude axes themselves. And don’t forget the time and level dimensions, they also belong to the metadata of
subdomain. -
If the coordinate bounds are not found exactly in the latitude/longitude coordinates, and accuracy around the map edges are of importance, you may need to do some 2D interpolation.
What about the Kelvin – Celsius conversion? The numerical conversion is easy:
var_nc4_c=var_nc4[:]-273.
But var_nc4_c now is a plain numpy.ma.core.MaskedArray, it lost all the metadata. A var_nc4_c.long_name would give you this error message:
AttributeError: 'MaskedArray' object has no attribute 'long_name'
Don’t even think about dimension coordinates, var_nc4_c has none of such things with it.
I think you start to see the pattern. Although come with the metadata from the NetCDF file, the variable read by the netcdf4 package does not support metadata-preserving operations, and the data values and metadata start to fall apart as soon as you start to do any meaningful computations/operations with the variable. I feel that this is going against the very philosophy of the NetCDF format: metadata is bundled with the data values when saved in files, but are separated when read into the memory, as if they are something totally irrelevant.
And this is not just metaphysical, it can cause troubles in your programming. I actually started off using CDAT, then learned a bit of netcdf4 years later, and it was a huge pain. I found myself having to maintain the metadata, particularly those dimension coordinates all the time. Say I have a function that accepts some 3D (time, latitude and longitude) input data as argument, and the output of the function is another 3D data variable with a different shape. With CDAT‘s TransientVariable, I could just pass in the TransientVariable to the function, query the dimension coordinates when I need to, or derive a new TransientVariable as the return value, which is also equipped with its own metadata.
When doing the same with netcdf4, I will have to pass in 3 more variables besides the data value itself (the time, latitude and longitude axes) to the function. Similarly, for every such data variable, there are 3 more return values (the updated time, latitude and longitude axes) that accompany the returned data value. The code becomes notably messier, and such maintenance work doesn’t really provide much added value, but adds extra burdens, and more lines of code to nurture bugs.
This is the greatest limitation I observed in the netcdf4 Python library. I have some limited experiences with the NetCDF’s Fortran and Matlab libraries, and they both seem to share the similar API and workflow, and therefore the same limitation, i.e. a very loose connection between data and metadata.
Out of frustration, I ended up writing a rudimentary custom class, following the similar interfaces of CDAT, to store the data read by netcdf4. I wouldn’t recommend you to do the same, it is not trivial to wrap the numpy.ndarray data type. Luckily, someone did this for us already. CDAT, iris and xarray are all more advanced tools that could make the manipulation of NetCDF data, and your life a lot easier.
There are quite a lot of power tools in the CDAT package, some of which I never used before, so there is a lot of content worth talking about. I will make a series of posts in the future to cover some aspects of CDAT that I found most useful in my previous work.